DIGITtally has been updated! See Details
TUTORIAL: Finding Genes of Interest DIGITtally
Click on any of the icons to learn more!
Click here to download example data
You start with a Drosophila melanogaster tissue, or set of tissues, of interest.
You will then choose...
You will then choose...

User-Supplied Genes
If you have a set of genes you're interested in, DIGITtally will rank them by how "interesting" they are in your tissues.
Genes of interest can be defined using our Gene List Builder
You will then choose...
DIGITtally-Defined Genes
Using FlyAtlas1 and/or FlyAtlas2 data, DIGITtally will create a list of genes which are Enriched in and Specific to your tissues

We will then use information from any/all of...
Click on a Data Source for a quick explanation!
name
info
| DataSource | Slice of Pie | Source Info |
|---|---|---|
| FlyAtlas1 | 11.1 | A microarray-based Drosophila melanogaster tissue-specific gene expression database, comprising 18 individual tissues |
| FlyAtlas2 | 11.1 | An RNA-seq based Drosophila melanogaster sex- and tissue-specific gene expression database, comprising 20 individual tissues |
| FlyCellAtlas | 11.1 | A single-cell RNA-seq based Drosophila melanogaster gene expression atlas. Contains information on 16 individual tissues, and over 250 individual cell types |
| FlyBase | 11.1 | The major repository for Drosophila melanogaster information, containing data from over 100000 individual research papers. |
| DIOPT | 11.1 | The most reliable ortholog ideantifier for model organisms, DIOPT aggregates scores from 17 individual ortholog-finding algorithms to confidently identify orthologs for individual genes |
| MozAtlas | 11.1 | A microarray-based Anopheles gambiae sex- and tissue-specific gene expression database, comprising 8 individual tissues |
| MozTubules | 11.1 | A microarray-based Anopheles gambiae sex-specific Malpighian tubule gene expression database, containing data from Male, Female, Larval and mixed Adult populations |
| Aegypti-Atlas | 11.1 | An RNA-seq based Aedes aegypti tissue-specific gene expression database, comprising 9 individual tissues, including an optional division of the mosquito gut into substructures |
| SilkDB | 11.1 | An RNA-seq based Bombyx mori life-stage and tissue-specific gene expression database, comprising 16 individual tissues. Notably, SilkDB contains expression data from 7 individual silkworm life stages |
TUTORIAL: Choosing Data Source Options

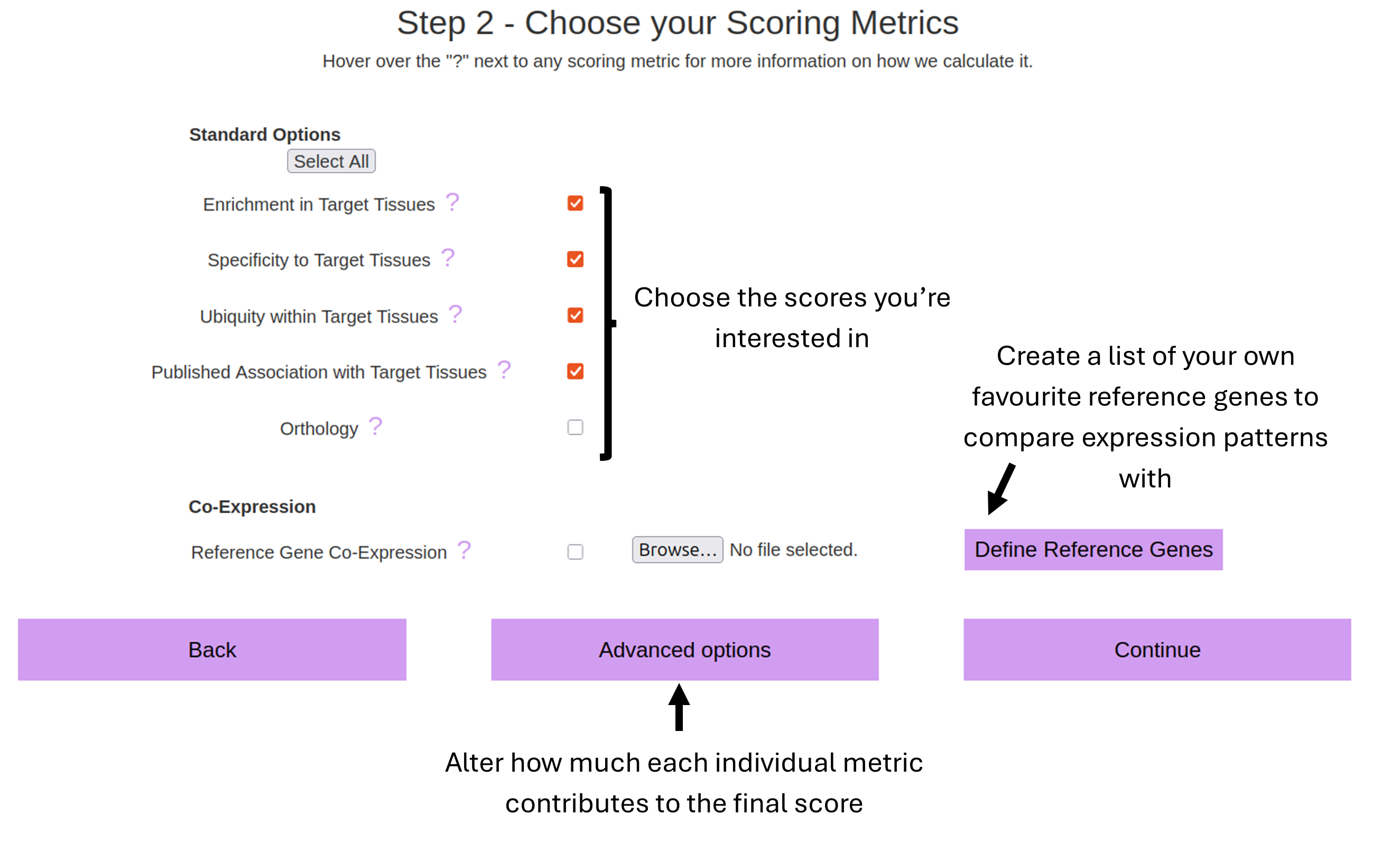
To score genes based on any/all of...
Click on a Data Source for a quick explanation!
| ScoreMetric | Slice of Pie | Score Info |
|---|---|---|
| Enrichment | 16.7 | Enrichment of each gene in tissues of interest compared to the whole fly |
| Specificity | 16.71 | Whether a gene appears to be specifically expressed or upregulated within the user-defined tissue(s) of interest. |
| Ubiquity | 16.7 | The extent to which a gene is expressed in all cells defined as cells of interest. |
| Co-expression | 16.7 | Co-expression of genes with a panel of user-submitted Reference Genes. |
| Published Association | 16.7 | Whether a gene has been demonstrated in the literature to have any role associated with the tissue(s) of interest. |
| Orthology | 16.7 | How well expression patterns in other insect species (A. gambiae, A. aegypti and B. mori) mirror those from Drosophila melanogaster within tissues of interest. Orthology scores can also take H. sapiens into account, ranking your genes by whether they have an "interesting" human ortholog. |
TUTORIAL: Choosing Scoring Metric Options
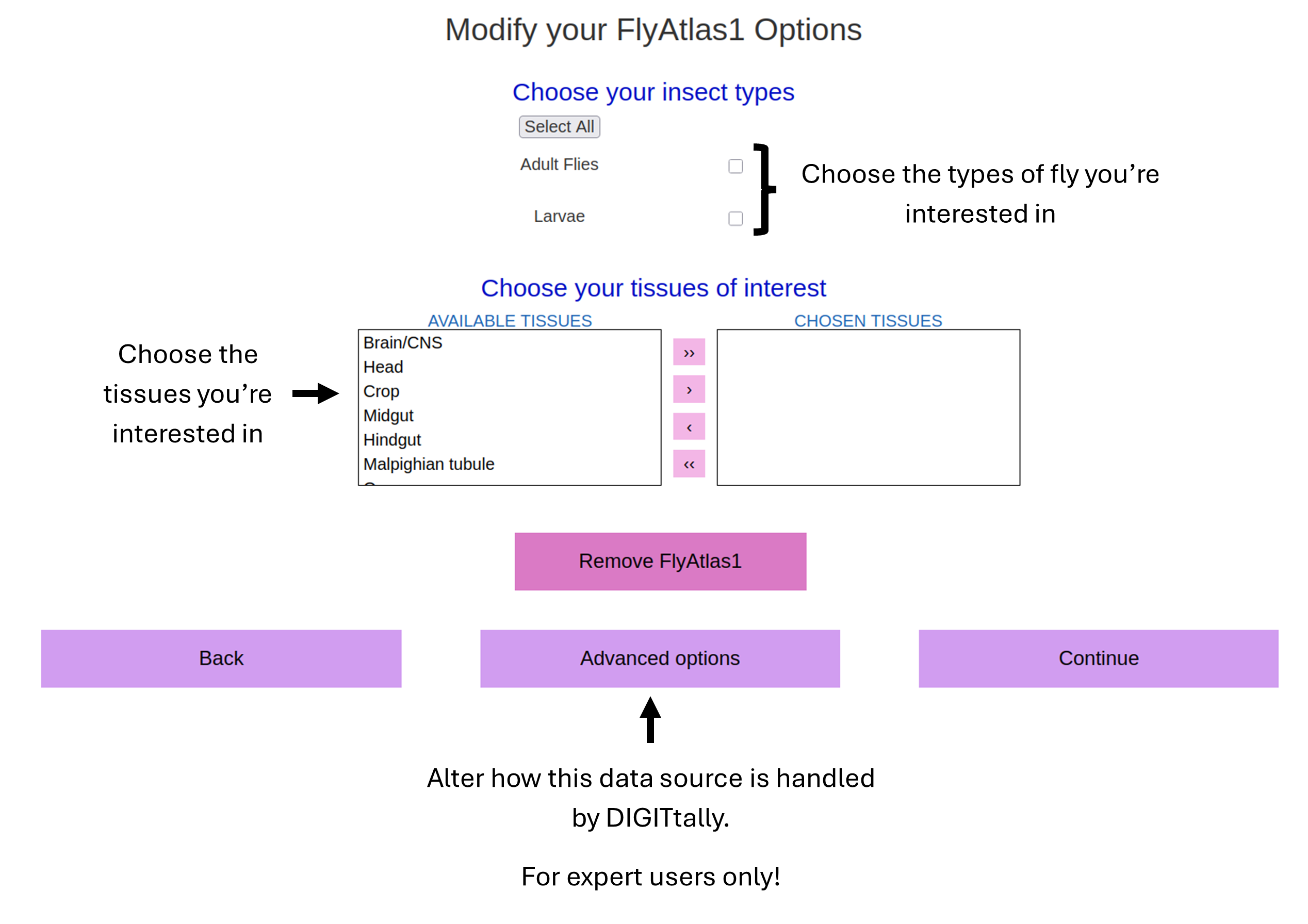
TUTORIAL: Choosing Individual Data Source Options (Example: FlyAtlas1)
...thus producing an individualised, ranked gene of interest list!
So why not try it out using one of the buttons below?
TUTORIAL: Finding Genes of Interest DIGITtally
Please note: For optimal user experience, the desktop version of DIGITtally is recommended
You start with a Drosophila melanogaster tissue, or set of tissues, of interest.
You will then choose...
Press one of the buttons above to learn about how DIGITtally handles Gene Lists, or use the button below to see the next step of the DIGITtally process
We will then use information from any/all of the following Data Sources. Choose one for a description!
Use the dropdown menu to learn more about our Data Sources!
Data from these sources will be used to score genes based on any/all of the following metrics. Choose one for a description!
Use the dropdown menu to learn more about our Scoring Metrics!
...thus producing an individualised, ranked gene of interest list!
So why not try it out using one of the buttons below, or explore the site via the sidebar?
DIGITtally will always be free and open to all users, no login required
To store your results, DIGITtally uses session cookies. No additional/external cookies are used.

